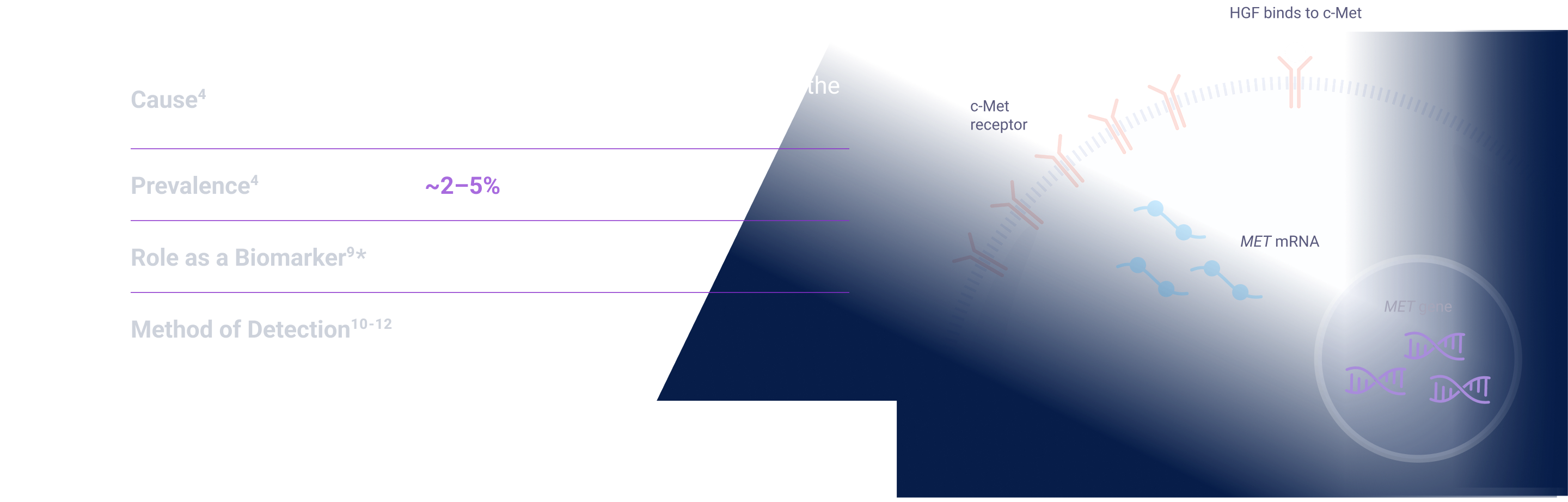
MET signaling in NSCLC is dysregulated through mechanisms including, but not limited to, high c-Met protein overexpression, MET gene amplification, METex14 skipping mutations, and HGF overexpression. MET aberrations have been linked to poor prognosis in NSCLC3-7
Depending on the testing methodology and patient population, MET aberration prevalence can be up to:
The prevalence of biomarkers does not indicate relative efficacy or safety of associated therapies. Some biomarkers may have greater characterization across various patient populations.
*Threshold for c-Met protein overexpression is ≥50% tumor cells with strong (3+) staining intensity. †EGFR-WT NSQ NSCLC.
EGFR=epidermal growth factor receptor; ex14=exon 14; HGF=hepatocyte growth factor; MET=mesenchymal-epithelial transition; NSCLC=non-small cell lung cancer; NSQ=non-squamous; WT=wild type.
*Threshold for c-Met protein overexpression is ≥50% tumor cells with strong (3+) staining intensity. †EGFR-WT NSQ NSCLC.
EGFR=epidermal growth factor receptor; ex14=exon 14; HGF=hepatocyte growth factor; IHC=immunohistochemistry; MET=mesenchymal-epithelial transition; NSCLC=non-small cell lung cancer; NSQ=non-squamous; WT=wild type.
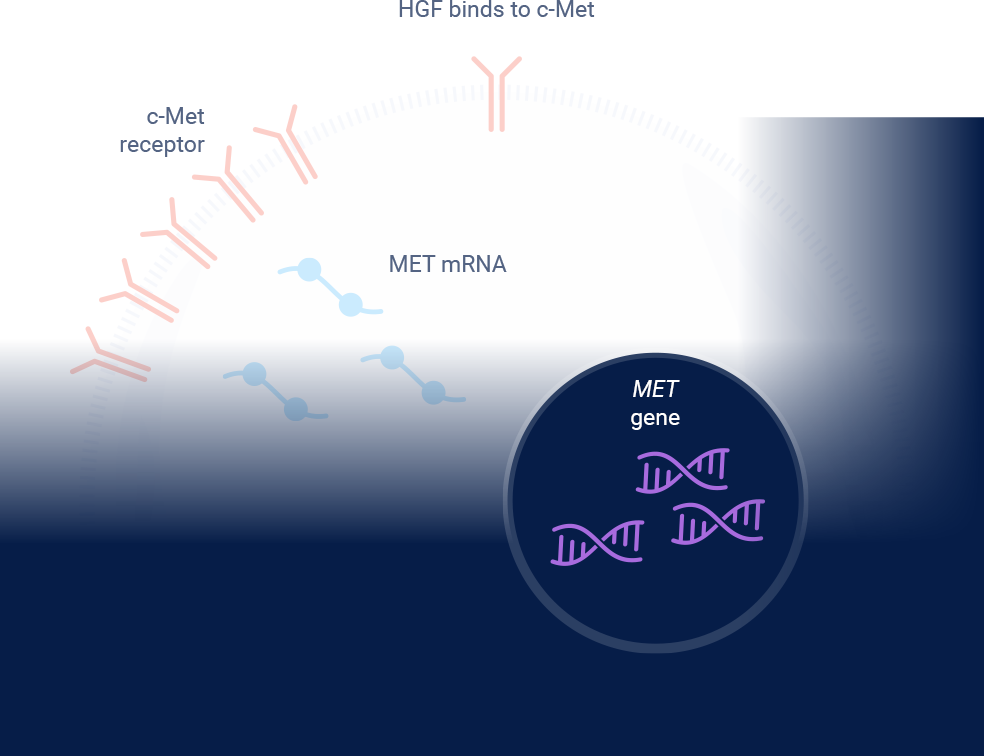
*Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Non-Small Cell Lung Cancer V.3.2025. © National Comprehensive Cancer Network, Inc. 2025. All rights reserved. Accessed March 31, 2025. To view the most recent and complete version of the guideline, go online to NCCN.org. NCCN makes no warranties of any kind whatsoever regarding their content, use or application and disclaims any responsibility for their application or use in any way.
ex14=exon 14; FISH=fluorescence in situ hybridization; HGF=hepatocyte growth factor; MET=mesenchymal-epithelial transition; NGS=next-generation sequencing; NSCLC=non-small cell lung cancer; RT-PCR=reverse transcription polymerase chain reaction.
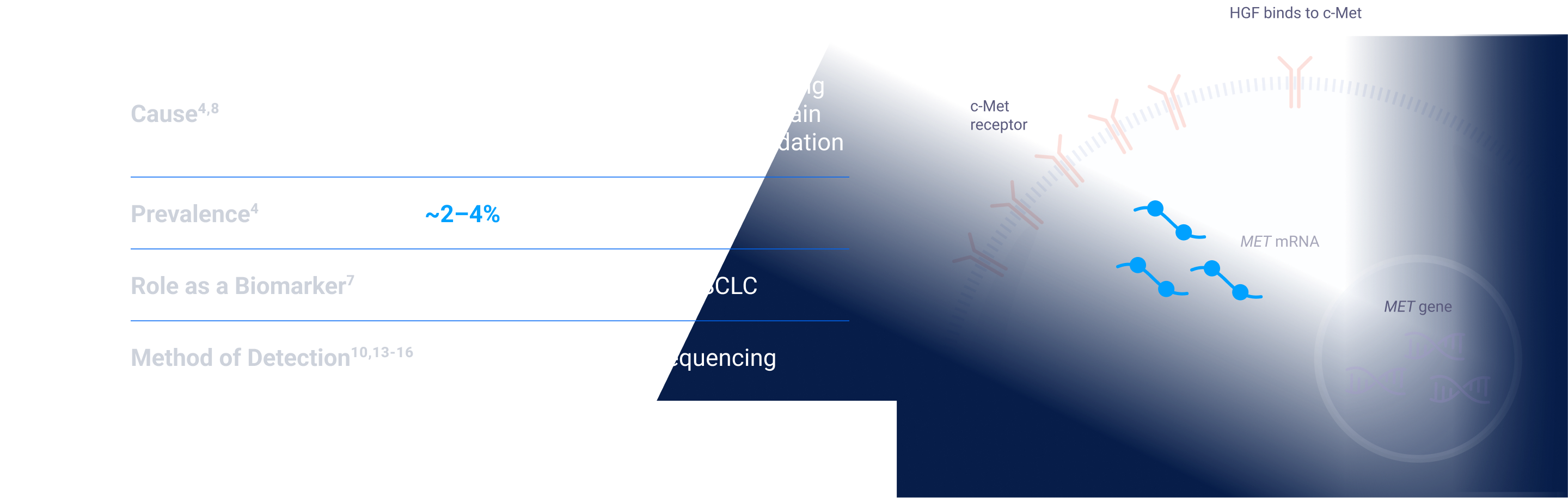
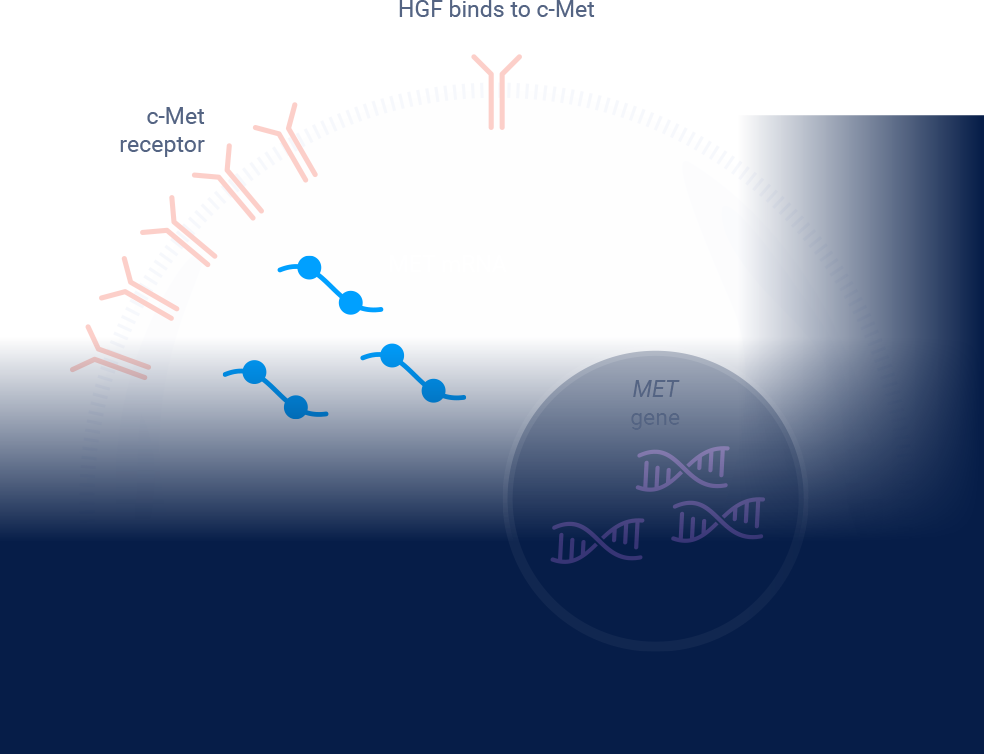
ex14=exon 14; HGF=hepatocyte growth factor; MET=mesenchymal-epithelial transition; NGS=next-generation sequencing; NSCLC=non-small cell lung cancer; RT-PCR=reverse transcription polymerase chain reaction.
MET amplification is an emerging biomarker and in clinical research as a potential therapeutic target. Prognosis statements and prevalence estimates are based on multiple sources; survival and prevalence data can vary among studies and datasets because of detection methodology used, patient sample sizes and/or demographics/characteristics. Some patients may have more than one MET aberration and may have overlap with other NSCLC biomarkers.